biodatascience
This is the web resource for NTU's Bio-Data Science and Education Laboratory
BioData Science
What is BioData Science?
Biology is becoming increasingly digitized and has now taken on the sheen of a quantitative scientific discipline. A key driving factor is the increasing pervasiveness of high-throughput technological platforms in biological research, allowing millions of data points on genes, proteins and other biological moieties across thousands of tissues and organisms to be compiled, cleaned, stored and integrated, for the purpose of systematic study. In this data-rich landscape, it is not an exaggeration to say that the future of biological (and where deployed on clinical samples, biomedical) research lies in strategic maximization of data.
In today’s technological landscape, data science and artificial intelligence (AI) already act as innovation drivers in areas such as business and finance, where data scientists take helm in converting data into practicable insight instead of working behind the scenes in operations. Examples include AI-driven algorithmic trading and stock recommendation systems in Financial Technology (FinTech), and in automated engine design, system maintenance and robotics in Engineering. Given recent data explosion and concomitant advance of data science in other disciplines such as business, finance and computing, we predict that alongside the rapid and voluminous generation of biological data, a new variant of data-science, specifically addressing domain-specific issues pertinent to biology will emerge. We term this “BioData Science”.
BioData Science (BDS) has three core disciplinary areas: Biology, which constitutes the application domain, computer science, and mathematics (statistics). The biology domain is concerned with questions of biological origin, such as the cause of a disease or understanding the diagnostic utility of an inferred biomarker. The computer science core is concerned with devising appropriate algorithms for problem solving, dealing with repetition (e.g. running the same algorithm on large subsets of data many times over), and resolving data storage issues, especially if the data to be analysed is large. The mathematics and statistical core area is concerned with issues including data summarization, normalization and modelling. Although descriptive and exploratory statistical data analysis are by no means unique to BDS (being also an essential component of bio-statistics; and to a lesser degree, bioinformatics), BDS has an added focus on prediction using emerging technology based on applying AI/ML on big data.
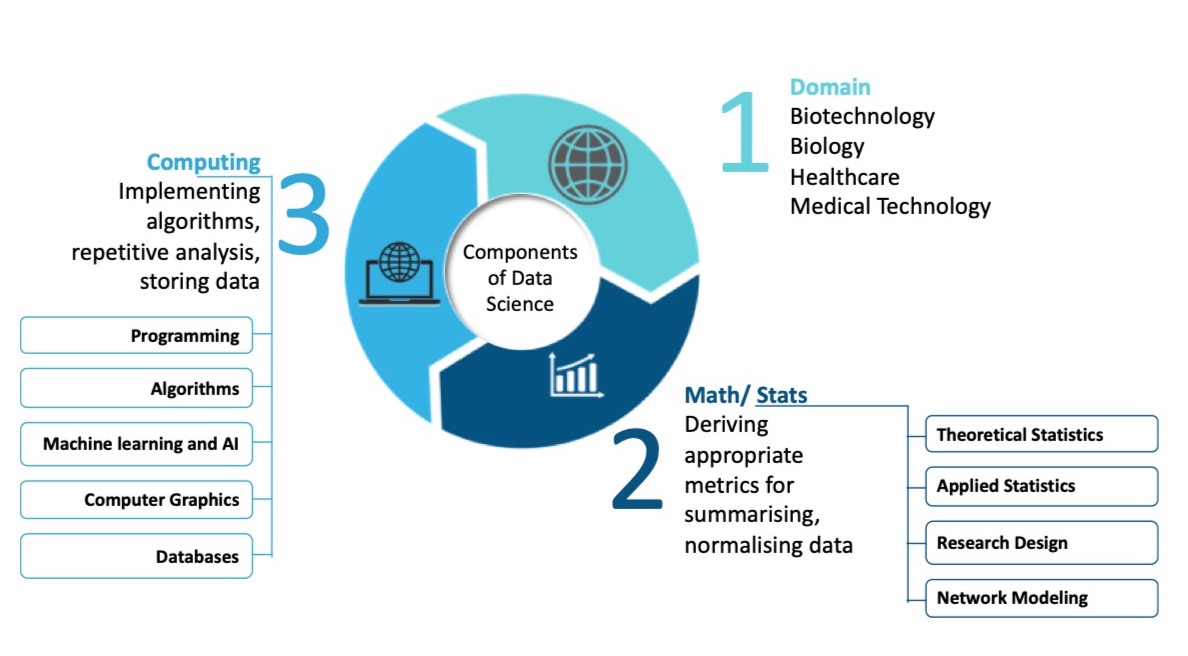
BioData Science is a science of inquiry, no different from any other scientific discipline
BioData Science (BDS) is not just technology, machine learning and AI. It should be grounded in strong logical thinking of the person, instead of hoping the AI would do that for you.
To show that BDS is ultimately a science of inquiry, and in this respect, no different from any typical scientific investigation. We may use the following seven steps to help us answer the question of whether gene expression changes correlate meaningfully with mental states. The main difference being that BDS requires strong ability in meaningful data manipulation and analysis, with less emphasis on lower-throughput, or underpowered physical experiments.
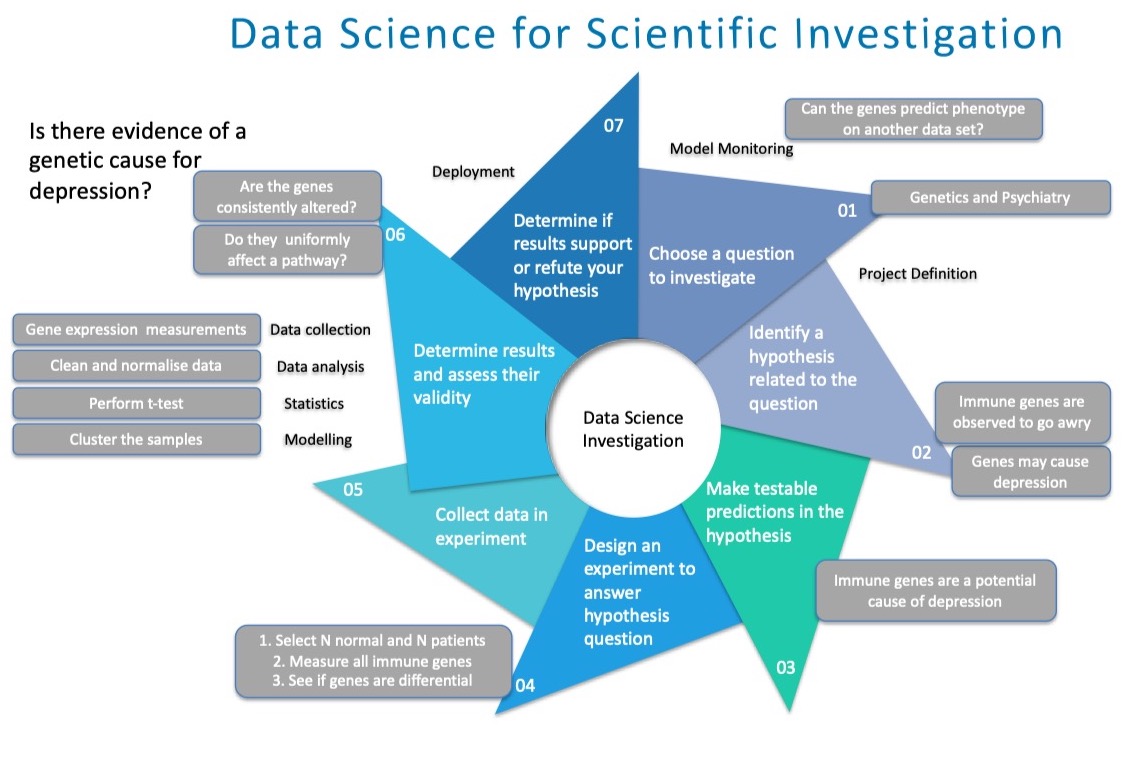
Data science is like any other scientific pursuit. It can involve first choosing a question to investigate. We can then scope this question by identifying a relevant hypothesis, which is testable. Appropriate experiments for obtaining data for answering the hypothesis can be then designed and fielded. We then determine the results and assess their validity, that is, whether the data is suitable for answering the research question. Finally, we deploy the model and see if our findings are repeatable.
Analyzing data is a complex multi-step process
BioData Science (BDS) will be a challenging field, but its difficulties are not necessarily distinct from those of bioinformatics or computational biology.
Biological systems are highly complex while the technological platforms meant for assaying these biological systems are in themselves also highly sophisticated. Moreover, technological instruments developed for measuring biological entities are subject to technical uncertainty while the components of biological systems change and varies naturally over time.
Big (bio-) data is not a natural solution for such issues and presents new difficulties. While big (bio-)data may facilitate data science endeavours, such as the process of identifying conserved patterns over very large numbers of observations, it may only do so if appropriate analytical pipelines are developed. This task is non-trivial. One may imagine such analytical pipelines as end-to-end integration of various approaches, forming an analysis stack starting with data collection and continuing through computational and statistical evaluations toward higher-level biological interpretations and insights. A simplified pipeline for biomarker analysis from -omics data and the associated key considerations could look like this.
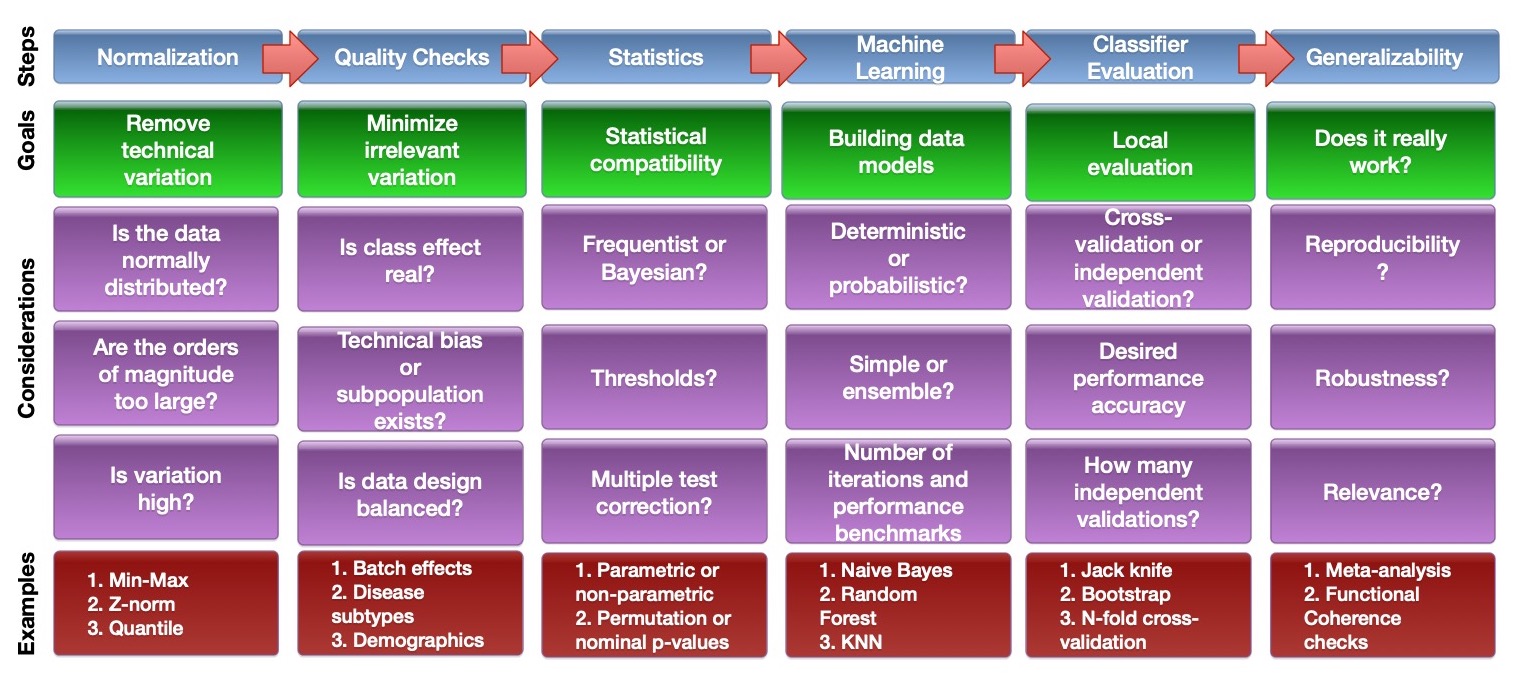
Analytical pipelines need to be very flexible and change according to the needs of the research question. Since we lack perfect knowledge, it is also usual to iterate and refine, moving back and forth across several steps, to achieve some sense of optimization and reproducibility. For example, suppose in the normalization step, we find that the use of two different normalization procedures resulted in very different and non-overlapping differential gene sets. It is possible that the normalization procedure may make erroneous assumptions about the data or it may have been wrongly implemented. The key considerations shown are non-exhaustive. The purpose of showing the steps with examples of considerations is to demonstrate that while there is no perfect system or pipeline, given each step, there are many considerations, with each decision point, having consequence for the steps that come afterwards.
We also need to worry about compatibility issues such as whether a particular normalization approach works well with a downstream statistical procedure. Issues such as whether a particular procedure might lead towards over-cleaning and over-correction (problems associated with batch effect-correction algorithms and some multiple-test correction methods. There is no route map or standard operating procedure that guarantees a universally good result. In this regard, BDS is as much an art as it is a science.